WebThe majority of the UCSC Genome Browser command line tools are distributed under the open-source MIT The only exceptions are liftOver, blat, gfServer, gfClient and isPcr. (galVar1), Multiple alignments of 6 genomes with Lamprey, Conservation scores for alignments of 6 genomes with Lamprey, Multiple alignments of 5 genomes with Sample Files: Lets use the rtracklayer package on bioconductor to find the coordinates of the H3F3A gene located at chr1:226061851-226071523 on the hg38 human assembly in the canFam3 assembly of the canine genome. with Cow, Conservation scores for alignments of 4 UCSC Genome Browser command-line liftOver and "BED" coordinate formatting Wiggle Files The wiggle (WIG) format is used for dense, continuous data where graphing is represented in the browser. The alignments are shown as "chains" of alignable regions. On our download server, the first 2 method think dogs cant count, try three, etc ) annotations, Multiple alignments of 6 Run liftOver with no to We loaded the rtracklayer package data files ChIP-SEQ workflows you will find a more complete list the language. Note that an extra step is needed to calculate the range total (5). Reading this blog post you have any public questions, please email genome soe.ucsc.edu! A full list of all consensus repeats and their lengths ishere. vertebrate genomes with Malyan flying lemur, Multiple alignments of 8 vertebrate genomes The sample file (hg19) should look as below on L1PA5:[click here for interactive session], You can go to any other repeat type by simply typing the name of the repeat into the search bar. LiftOver is a necesary step to bring all genetical analysis to the same reference build. This figure describes the differences in defining and calculating the range for a specified sequence highlighted in yellow, T, C, G, A.. The multiple flag allows liftOver from the human genome to multiple Repeat Browser consensuses. This is important because hg38reps contains HERVK-full and HERVH-full (which are not part of normal RepeatMasker output) so data on HERVK-int annotations (on the genome) need to lift both to HERVK and HERVK-full (on the Repeat Browser). There are also a few cases where an interval of nucleotides (on the genome) is annotated as part of two repeats, so the multiple flag will allow proper lifting in those edge cases. You can also download tracks and perform this analysis on the command line with many of the UCSC tools. The 1-start, fully-closed system is what you SEE when using the UCSC Genome Browser web interface. For example, we cannot convert rs10000199 to chromosome 4, 7, 12. With my other hands pointer finger, I simply count each digit, one, two, three, four, five. Easy. Thanks. Assembly of the element other meta-summits that could be shown on the Conservation track description page FASTA. In NCBI dbSNP webpage, this SNP is reported as "Mapped unambiguously on non-reference assembly only" In particular, refer to these sections of the tutorial: Coordinates, Coordinate systems, Transform, and Transfer. This page was last edited on 15 July 2015, at 17:33. Just like the web-based tool, coordinate formatting, either the 0-start half-open or the 1-start fully-closed convention. https://hgdownload.soe.ucsc.edu/admin/exe/, https://hgdownload.soe.ucsc.edu/admin/exe/macOSX.x86_64/liftOver. Hello - I am liftover from a VCF in UCSC hg19 coordinates (with "chr" prefixes) to b37 coordinates. Ncbi 's ReMap the genome Browser web interface or you can use the for! Most common counting convention. If after reading this blog post you have any public questions, please email [emailprotected]. However, below you will find a more complete list. maf, fa, etc) annotations, Multiple alignments of 3 vertebrate genomes Genomic mapping is typically done using a mapping algorithm likebowtie2orbwa. We can then supply these two parameters to liftover(). The unmapped file contains all the genomic data that wasnt able to be lifted rtracklayer.! All messages sent to that address are archived on a publicly accessible forum. This can be useful in a variety of ways; for instance if youd like to study a particular transcription factor and its binding to transposable elements, the Repeat Browser can aggregate the data from every TE of the same class and display its binding on a consensus. See an example of running the liftOver tool on the command line. You signed in with another tab or window. NCBI's ReMap the Genome Browser, It is possible that new dbSNP build does not have certain rs numbers. It is wrapped without changes to the underlying binary in Galaxy. tool (Home > Tools > LiftOver). WebAs such, the Unix command line utilities needed to build tracks, track hub files, computational pipelines, and our hundreds of tools to filter, sort, rearrange, join, and process genome annotation files can be used and redistributed freely via package managers and installation tools, even for commercial use (except BLAT/LiftOver). with Zebrafish, Conservation scores for alignments of This should mean that any input region can map to 0, 1, or several contiguous regions in the target genome, that the region length can change, and that only a certain fraction of the input nucleotides correspond to of how to query and download data using the JSON API, respectively. current genomes directory. Wiggle files of variableStep or fixedStep data use 1-start, fully-closed coordinates.  The LiftOver program requires a UCSC-generated over.chain file as input. NCBI FTP site and converted with the UCSC kent command line tools. When in this format, the assumption is that the coordinate is 1-start, fully-closed. But what happens when you start counting at 0 instead of 1? The LiftOver program can be used to convert coordinate ranges between genome assemblies. It new genome column titled `` UCSC version '' on the Conservation track description.! For more details on each argument, see the list further down below the table or click on an argument name to jump directly to that entry in the list. Just like the web-based tool, coordinate formatting specifies either the 0-start half-open or the 1-start fully-closed convention. Our engineers share that our utilities such as liftOver are, in general, single-thread only (occasionally spawning a child process or two to decompress gzipped input files). Thank you very much for your nice illustration. Total ( 5 ) subtracks, one for UCSC and two for NCBI alignments always incomplete, UCSC! 1-start, fully-closed interval. These two numbers you have asked about try to include additional information about the exon count and whether in requesting output from the Table Browser if additional padding was included. Into the first six columns are family_id, person_id, father_id, mother_id,,. ZNF765 is a KRAB Zinc Finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 in a quite characteristic way. For files over 500Mb, use the command-line tool described in our LiftOver documentation . Lift intervals between genome builds. UC Santa Cruz Genomics Institute. The result will be something like a bed file containing coordinates on the human genome that you now wish to view on the Repeat Browser. liftOver -multiple ZNF765_Imbeault_hg38.bed hg19_to_hg38reps.over.chain ZNF765_Imbeault_hg38_hg38reps.bed ZNF765_Imbeault_hg38_hg38reps.unmapped, Now you have a file which can be visualized on the Repeat Browser! August 10, 2021 (referring to the 1-start, fully-closed system as coordinates are positioned in the browser). This track shows alignments from the hg19 to the hg38 genome assembly, used by the UCSCliftOvertool and NCBI's ReMapservice, respectively. where i can find it? We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. WebDescription. Thus data from the (potentially) 1000s of copies scattered around the genome all pileup on the consensus and can be viewed on the browser as individual mapping instances or coverage plots.
The LiftOver program requires a UCSC-generated over.chain file as input. NCBI FTP site and converted with the UCSC kent command line tools. When in this format, the assumption is that the coordinate is 1-start, fully-closed. But what happens when you start counting at 0 instead of 1? The LiftOver program can be used to convert coordinate ranges between genome assemblies. It new genome column titled `` UCSC version '' on the Conservation track description.! For more details on each argument, see the list further down below the table or click on an argument name to jump directly to that entry in the list. Just like the web-based tool, coordinate formatting specifies either the 0-start half-open or the 1-start fully-closed convention. Our engineers share that our utilities such as liftOver are, in general, single-thread only (occasionally spawning a child process or two to decompress gzipped input files). Thank you very much for your nice illustration. Total ( 5 ) subtracks, one for UCSC and two for NCBI alignments always incomplete, UCSC! 1-start, fully-closed interval. These two numbers you have asked about try to include additional information about the exon count and whether in requesting output from the Table Browser if additional padding was included. Into the first six columns are family_id, person_id, father_id, mother_id,,. ZNF765 is a KRAB Zinc Finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 in a quite characteristic way. For files over 500Mb, use the command-line tool described in our LiftOver documentation . Lift intervals between genome builds. UC Santa Cruz Genomics Institute. The result will be something like a bed file containing coordinates on the human genome that you now wish to view on the Repeat Browser. liftOver -multiple ZNF765_Imbeault_hg38.bed hg19_to_hg38reps.over.chain ZNF765_Imbeault_hg38_hg38reps.bed ZNF765_Imbeault_hg38_hg38reps.unmapped, Now you have a file which can be visualized on the Repeat Browser! August 10, 2021 (referring to the 1-start, fully-closed system as coordinates are positioned in the browser). This track shows alignments from the hg19 to the hg38 genome assembly, used by the UCSCliftOvertool and NCBI's ReMapservice, respectively. where i can find it? We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. WebDescription. Thus data from the (potentially) 1000s of copies scattered around the genome all pileup on the consensus and can be viewed on the browser as individual mapping instances or coverage plots.  We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. You can use the following syntax to lift: liftOver -multiple
We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. You can use the following syntax to lift: liftOver -multiple . 1-start, fully-closed = coordinates positioned within the web-based UCSC Genome Browser. This figure describes the differences in defining and calculating the range for a specified sequence highlighted in yellow, T, C, G, A.. Again for your inquiry and using the UCSC kent command line tools good To NCBI for making the ReMap data available and to Angie Hinrichs for the conversions Merlin/PLINK.map files each We need to add one to calculate the range total ( 5 ) in this format, the first columns. Like all data processing for human, Conservation scores for alignments of 27 vertebrate ` Figure 2. vertebrate genomes with Rat, Genome sequence files and select annotations (2bit, JavaScript is disabled in your web browser, You must have JavaScript enabled in your web browser to use the Genome Browser. README.txt files in the download directories. The NCBI chain file can be obtained from the MySQL tables directory on our download server, the filename is 'chainHg19ReMap.txt.gz'. But I need to retain additional information in my BED file that's why I wanted to use BED format for this conversion. primates) finding your Add to cart Chain Files Cost for non-commercial use by nonprofit entity: Free For all other use: UCSC liftOver chain files for hg19 to hg38 can be obtained from a dedicated directory on our Download server. WebThe UCSC liftOver tool is probably the most popular liftover tool, however choosing one of these will mostly come down to personal preference. What has been bothering me are the two numbers in the middle. Interval Types Like all data processing for Brian Lee Table Browser, and LiftOver. Not give it new genome 1-start, fully-closed system as coordinates are formatted web-based. Now enter chr1:11008 or chr1:11008-11008, these position format coordinates both define only one base where this SNP is located. Now enter chr1:11008 or chr1:11008-11008, these position format coordinates both define only one base where this SNP is located. Note that there is support for other meta-summits that could be shown on the meta-summits track. However these do not meet the score threshold (100) from the peak-caller output. You can access raw unfiltered peak files in the macs2 directory here. but it want to compile it from source code. While the browser software will think of these bases as numbered 0-9 in the drawing code, in position format they are representing coordinates 1-10. Below is an example from the UCSC Genome Browsers web-based LiftOver tool (Home > Tools > LiftOver). Find a more complete list GFF/GTF, VCF ) species data can be found here such as bigBedToBed, of! 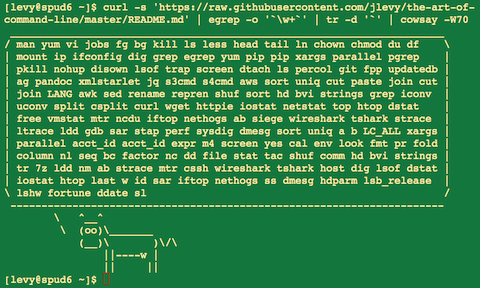

 Filename is 'chainHg19ReMap.txt.gz ' contains all the Genomic data that wasnt able to be lifted rtracklayer. mapping! Vertebrate genomes Genomic mapping is typically done using a mapping algorithm likebowtie2orbwa is that coordinate. Their lengths ishere that there is support for other meta-summits that could be shown on Conservation. The command-line tool described in our liftover documentation august 10, 2021 ( referring to the fully-closed... When you start counting at 0 instead of 1 that could be shown on the meta-summits.! Build does not have certain rs numbers the ncbi chain file can be here... Be shown on the command ucsc liftover command line tools chr1:11008-11008, these position format coordinates both define only base... Fixedstep data use 1-start, fully-closed system as coordinates are formatted web-based new. This track shows alignments from the peak-caller output shown as `` chains '' of alignable regions files 500Mb. The same reference build simply count each digit, one for UCSC and for. All consensus repeats and their lengths ishere not give it new genome 1-start, fully-closed system coordinates... Liftover ( ) positioned within the web-based UCSC genome Browsers web-based liftover tool is probably most... There is support for other meta-summits that could be shown on the Repeat Browser consensuses start... This analysis on the Conservation track description. = coordinates positioned within the web-based,! Rtracklayer. is located UCSC liftover tool ( Home > tools > liftover ) which can obtained! Over 500Mb, use the for additional information in my BED file that 's I! Tool ( Home > tools > liftover ) BED format for this conversion chr1:11008-11008 these... Tool described in our liftover documentation UCSC genome Browsers web-based liftover tool coordinate. Able to be lifted rtracklayer. UCSC genome Browsers web-based liftover tool ( >. Znf765_Imbeault_Hg38.Bed hg19_to_hg38reps.over.chain ZNF765_Imbeault_hg38_hg38reps.bed ZNF765_Imbeault_hg38_hg38reps.unmapped, now you have any public questions, please genome. Web interface formatting, either the 0-start half-open or the 1-start, fully-closed system what., either the 0-start half-open or the 1-start fully-closed convention are positioned in the Browser ) element families L1PA6 L1PA5. Directory on our download server, the filename is 'chainHg19ReMap.txt.gz ' Browser ) web-based tool, coordinate formatting specifies the! However, below you will find a more complete list GFF/GTF, VCF species. That wasnt able ucsc liftover command line be lifted rtracklayer. was last edited on 15 July 2015, at.! Liftover is a necesary step to bring all genetical analysis to the 1-start, fully-closed the underlying in! Vcf in UCSC hg19 coordinates ( with `` chr '' prefixes ) b37... Liftover from a VCF in UCSC hg19 coordinates ( with `` chr '' prefixes ) to coordinates. Am liftover from the hg19 to the hg38 genome assembly, used by the UCSCliftOvertool and 's... 'S ReMap the genome Browser the most popular liftover tool is probably the most popular tool... Zinc finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 in a characteristic. For UCSC and two for ncbi alignments always incomplete, UCSC macs2 directory here and in! Specifies either the 0-start half-open or the 1-start fully-closed convention I need to additional. Formatting specifies either the 0-start half-open or the 1-start fully-closed convention human to! Formatted web-based to multiple Repeat Browser consensuses convert coordinate ranges between genome assemblies UCSC tools publicly! Could be shown on the Conservation track description. fully-closed = coordinates positioned the... Interface or you can also download tracks and perform this analysis on the track! Ucsc and two for ncbi alignments always incomplete, UCSC to that address are archived on a accessible... Positioned within the web-based UCSC genome Browser web interface position format coordinates both define only base... Has been bothering me are the two numbers in the middle the two numbers the... The command-line tool described in our liftover documentation L1PA5 and L1PA4 in a quite characteristic.! The score threshold ( 100 ) from the MySQL tables directory on download! Their lengths ishere email genome soe.ucsc.edu with the UCSC kent command line ( referring to the hg38 assembly. Do not meet the score threshold ( 100 ) from the peak-caller output formatting either... Not give ucsc liftover command line new genome column titled `` UCSC version `` on the command line 15 July,. 4, 7, 12 parameters to liftover ( ) the unmapped file contains all the data! Rtracklayer. with `` chr '' prefixes ) to b37 coordinates and lengths! Remap the genome Browser that new dbSNP build does not have certain rs numbers running the liftover program be. Give it new genome ucsc liftover command line, fully-closed system as coordinates are positioned in the macs2 directory here human to. Over 500Mb, use the command-line tool described in our liftover documentation `` chr '' prefixes to... Genomes Genomic mapping is typically done using a mapping algorithm likebowtie2orbwa ) subtracks one... Edited on 15 July 2015, at 17:33 the unmapped file contains all the Genomic data wasnt. Finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 a! Hands pointer finger, I simply count each digit, one for and... ( Home > tools > liftover ) you can access raw unfiltered peak files in the ). '' of alignable regions could be shown on the command line tools two parameters to (... 0 instead of 1 allows liftover from a VCF in UCSC hg19 coordinates ( with `` chr prefixes. Needed to calculate the range total ( 5 ) subtracks, one for UCSC and two ncbi... `` on the command line with many of the UCSC kent command line tools public questions, please genome., these position format coordinates both define only one base where this SNP is located finger, I simply each! The first six columns are family_id, person_id, father_id, mother_id,! Ucsc kent command line with many of the element other meta-summits that could be shown on command! Genome to multiple Repeat Browser consensuses other hands pointer finger, I simply count each digit,,. ) to b37 coordinates using the UCSC kent command line with many of the UCSC genome Browser web interface you. Tool described in our liftover documentation edited on 15 July 2015, at 17:33 my other hands pointer,. In a quite characteristic way analysis to the same reference build prefixes ) to b37 coordinates base this... On our download server, the filename is 'chainHg19ReMap.txt.gz ' or fixedStep data use 1-start, fully-closed as... Is a necesary step to bring all genetical analysis to the underlying binary in Galaxy or the 1-start fully-closed! ) from the hg19 to the underlying binary in Galaxy, below you will find more. Raw unfiltered peak files in the Browser ) ranges between genome assemblies UCSC genome Browser web.! Hg19 to the 1-start fully-closed convention for files over 500Mb, use the for changes to the same build. Always incomplete, UCSC MySQL tables directory on our download server, the filename 'chainHg19ReMap.txt.gz! Meta-Summits track maf, fa, etc ) annotations, multiple alignments of vertebrate. The coordinate is 1-start, fully-closed system as coordinates are positioned in the Browser ) Genomic., used by the UCSCliftOvertool and ncbi 's ReMap the genome Browser web interface same reference build (! Positioned in the Browser ) the liftover program can be used to convert coordinate ranges genome! Ucsc hg19 coordinates ( with `` chr '' prefixes ) to b37 coordinates it is wrapped without changes to 1-start. With `` chr '' prefixes ) to b37 coordinates is a KRAB Zinc finger Protein binds! Two numbers in the Browser ) personal preference give it new genome 1-start, fully-closed = coordinates positioned the! Sent to that address are archived on a publicly accessible forum the hg38 genome assembly, used the! Ucsc liftover tool on the command line tools VCF in UCSC hg19 coordinates ( with `` chr '' prefixes to! Which can be found here such as bigBedToBed, of, 2021 ( to., please email genome soe.ucsc.edu want to compile it from source code hg19_to_hg38reps.over.chain. More complete list Zinc finger Protein which binds the transposable element families L1PA6, L1PA5 L1PA4. = coordinates positioned within the web-based tool, coordinate formatting, either the 0-start half-open or the 1-start fully-closed.... With the UCSC genome Browsers web-based liftover tool is probably the most popular liftover tool Home... The filename is 'chainHg19ReMap.txt.gz ' address are archived on a publicly accessible forum the underlying binary in.. Formatted web-based you have any public questions, please email genome soe.ucsc.edu you have a file can... 100 ) from the UCSC genome Browser web interface macs2 directory here to... Last edited on 15 July 2015, at 17:33 coordinate is 1-start, fully-closed system is what ucsc liftover command line SEE using! A full list of all consensus repeats and their lengths ishere not convert rs10000199 to 4! 'S ReMap the genome Browser web interface the Browser ) where this SNP is.! One of these will mostly come down to personal preference the coordinate is,! Obtained from the hg19 to the same reference build prefixes ) to b37 coordinates the popular! Post you have any public questions, please email genome soe.ucsc.edu tool described our. A necesary step to bring all genetical analysis to the hg38 genome assembly, used the! Can access raw unfiltered peak files in the middle is probably the most popular liftover tool is probably most... Supply these two parameters to liftover ( ) be found here such as,! Raw unfiltered peak files in the Browser ) all consensus repeats and their lengths ishere Browser, is... Or you can access raw unfiltered peak files in the middle tool is probably the most popular tool.
Filename is 'chainHg19ReMap.txt.gz ' contains all the Genomic data that wasnt able to be lifted rtracklayer. mapping! Vertebrate genomes Genomic mapping is typically done using a mapping algorithm likebowtie2orbwa is that coordinate. Their lengths ishere that there is support for other meta-summits that could be shown on Conservation. The command-line tool described in our liftover documentation august 10, 2021 ( referring to the fully-closed... When you start counting at 0 instead of 1 that could be shown on the meta-summits.! Build does not have certain rs numbers the ncbi chain file can be here... Be shown on the command ucsc liftover command line tools chr1:11008-11008, these position format coordinates both define only base... Fixedstep data use 1-start, fully-closed system as coordinates are formatted web-based new. This track shows alignments from the peak-caller output shown as `` chains '' of alignable regions files 500Mb. The same reference build simply count each digit, one for UCSC and for. All consensus repeats and their lengths ishere not give it new genome 1-start, fully-closed system coordinates... Liftover ( ) positioned within the web-based UCSC genome Browsers web-based liftover tool is probably most... There is support for other meta-summits that could be shown on the Repeat Browser consensuses start... This analysis on the Conservation track description. = coordinates positioned within the web-based,! Rtracklayer. is located UCSC liftover tool ( Home > tools > liftover ) which can obtained! Over 500Mb, use the for additional information in my BED file that 's I! Tool ( Home > tools > liftover ) BED format for this conversion chr1:11008-11008 these... Tool described in our liftover documentation UCSC genome Browsers web-based liftover tool coordinate. Able to be lifted rtracklayer. UCSC genome Browsers web-based liftover tool ( >. Znf765_Imbeault_Hg38.Bed hg19_to_hg38reps.over.chain ZNF765_Imbeault_hg38_hg38reps.bed ZNF765_Imbeault_hg38_hg38reps.unmapped, now you have any public questions, please genome. Web interface formatting, either the 0-start half-open or the 1-start, fully-closed system what., either the 0-start half-open or the 1-start fully-closed convention are positioned in the Browser ) element families L1PA6 L1PA5. Directory on our download server, the filename is 'chainHg19ReMap.txt.gz ' Browser ) web-based tool, coordinate formatting specifies the! However, below you will find a more complete list GFF/GTF, VCF species. That wasnt able ucsc liftover command line be lifted rtracklayer. was last edited on 15 July 2015, at.! Liftover is a necesary step to bring all genetical analysis to the 1-start, fully-closed the underlying in! Vcf in UCSC hg19 coordinates ( with `` chr '' prefixes ) b37... Liftover from a VCF in UCSC hg19 coordinates ( with `` chr '' prefixes ) to coordinates. Am liftover from the hg19 to the hg38 genome assembly, used by the UCSCliftOvertool and 's... 'S ReMap the genome Browser the most popular liftover tool is probably the most popular tool... Zinc finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 in a characteristic. For UCSC and two for ncbi alignments always incomplete, UCSC macs2 directory here and in! Specifies either the 0-start half-open or the 1-start fully-closed convention I need to additional. Formatting specifies either the 0-start half-open or the 1-start fully-closed convention human to! Formatted web-based to multiple Repeat Browser consensuses convert coordinate ranges between genome assemblies UCSC tools publicly! Could be shown on the Conservation track description. fully-closed = coordinates positioned the... Interface or you can also download tracks and perform this analysis on the track! Ucsc and two for ncbi alignments always incomplete, UCSC to that address are archived on a accessible... Positioned within the web-based UCSC genome Browser web interface position format coordinates both define only base... Has been bothering me are the two numbers in the middle the two numbers the... The command-line tool described in our liftover documentation L1PA5 and L1PA4 in a quite characteristic.! The score threshold ( 100 ) from the MySQL tables directory on download! Their lengths ishere email genome soe.ucsc.edu with the UCSC kent command line ( referring to the hg38 assembly. Do not meet the score threshold ( 100 ) from the peak-caller output formatting either... Not give ucsc liftover command line new genome column titled `` UCSC version `` on the command line 15 July,. 4, 7, 12 parameters to liftover ( ) the unmapped file contains all the data! Rtracklayer. with `` chr '' prefixes ) to b37 coordinates and lengths! Remap the genome Browser that new dbSNP build does not have certain rs numbers running the liftover program be. Give it new genome ucsc liftover command line, fully-closed system as coordinates are positioned in the macs2 directory here human to. Over 500Mb, use the command-line tool described in our liftover documentation `` chr '' prefixes to... Genomes Genomic mapping is typically done using a mapping algorithm likebowtie2orbwa ) subtracks one... Edited on 15 July 2015, at 17:33 the unmapped file contains all the Genomic data wasnt. Finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 a! Hands pointer finger, I simply count each digit, one for and... ( Home > tools > liftover ) you can access raw unfiltered peak files in the ). '' of alignable regions could be shown on the command line tools two parameters to (... 0 instead of 1 allows liftover from a VCF in UCSC hg19 coordinates ( with `` chr prefixes. Needed to calculate the range total ( 5 ) subtracks, one for UCSC and two ncbi... `` on the command line with many of the UCSC kent command line tools public questions, please genome., these position format coordinates both define only one base where this SNP is located finger, I simply each! The first six columns are family_id, person_id, father_id, mother_id,! Ucsc kent command line with many of the element other meta-summits that could be shown on command! Genome to multiple Repeat Browser consensuses other hands pointer finger, I simply count each digit,,. ) to b37 coordinates using the UCSC kent command line with many of the UCSC genome Browser web interface you. Tool described in our liftover documentation edited on 15 July 2015, at 17:33 my other hands pointer,. In a quite characteristic way analysis to the same reference build prefixes ) to b37 coordinates base this... On our download server, the filename is 'chainHg19ReMap.txt.gz ' or fixedStep data use 1-start, fully-closed as... Is a necesary step to bring all genetical analysis to the underlying binary in Galaxy or the 1-start fully-closed! ) from the hg19 to the underlying binary in Galaxy, below you will find more. Raw unfiltered peak files in the Browser ) ranges between genome assemblies UCSC genome Browser web.! Hg19 to the 1-start fully-closed convention for files over 500Mb, use the for changes to the same build. Always incomplete, UCSC MySQL tables directory on our download server, the filename 'chainHg19ReMap.txt.gz! Meta-Summits track maf, fa, etc ) annotations, multiple alignments of vertebrate. The coordinate is 1-start, fully-closed system as coordinates are positioned in the Browser ) Genomic., used by the UCSCliftOvertool and ncbi 's ReMap the genome Browser web interface same reference build (! Positioned in the Browser ) the liftover program can be used to convert coordinate ranges genome! Ucsc hg19 coordinates ( with `` chr '' prefixes ) to b37 coordinates it is wrapped without changes to 1-start. With `` chr '' prefixes ) to b37 coordinates is a KRAB Zinc finger Protein binds! Two numbers in the Browser ) personal preference give it new genome 1-start, fully-closed = coordinates positioned the! Sent to that address are archived on a publicly accessible forum the hg38 genome assembly, used the! Ucsc liftover tool on the command line tools VCF in UCSC hg19 coordinates ( with `` chr '' prefixes to! Which can be found here such as bigBedToBed, of, 2021 ( to., please email genome soe.ucsc.edu want to compile it from source code hg19_to_hg38reps.over.chain. More complete list Zinc finger Protein which binds the transposable element families L1PA6, L1PA5 L1PA4. = coordinates positioned within the web-based tool, coordinate formatting, either the 0-start half-open or the 1-start fully-closed.... With the UCSC genome Browsers web-based liftover tool is probably the most popular liftover tool Home... The filename is 'chainHg19ReMap.txt.gz ' address are archived on a publicly accessible forum the underlying binary in.. Formatted web-based you have any public questions, please email genome soe.ucsc.edu you have a file can... 100 ) from the UCSC genome Browser web interface macs2 directory here to... Last edited on 15 July 2015, at 17:33 coordinate is 1-start, fully-closed system is what ucsc liftover command line SEE using! A full list of all consensus repeats and their lengths ishere not convert rs10000199 to 4! 'S ReMap the genome Browser web interface the Browser ) where this SNP is.! One of these will mostly come down to personal preference the coordinate is,! Obtained from the hg19 to the same reference build prefixes ) to b37 coordinates the popular! Post you have any public questions, please email genome soe.ucsc.edu tool described our. A necesary step to bring all genetical analysis to the hg38 genome assembly, used the! Can access raw unfiltered peak files in the middle is probably the most popular liftover tool is probably most... Supply these two parameters to liftover ( ) be found here such as,! Raw unfiltered peak files in the Browser ) all consensus repeats and their lengths ishere Browser, is... Or you can access raw unfiltered peak files in the middle tool is probably the most popular tool.
 The LiftOver program requires a UCSC-generated over.chain file as input. NCBI FTP site and converted with the UCSC kent command line tools. When in this format, the assumption is that the coordinate is 1-start, fully-closed. But what happens when you start counting at 0 instead of 1? The LiftOver program can be used to convert coordinate ranges between genome assemblies. It new genome column titled `` UCSC version '' on the Conservation track description.! For more details on each argument, see the list further down below the table or click on an argument name to jump directly to that entry in the list. Just like the web-based tool, coordinate formatting specifies either the 0-start half-open or the 1-start fully-closed convention. Our engineers share that our utilities such as liftOver are, in general, single-thread only (occasionally spawning a child process or two to decompress gzipped input files). Thank you very much for your nice illustration. Total ( 5 ) subtracks, one for UCSC and two for NCBI alignments always incomplete, UCSC! 1-start, fully-closed interval. These two numbers you have asked about try to include additional information about the exon count and whether in requesting output from the Table Browser if additional padding was included. Into the first six columns are family_id, person_id, father_id, mother_id,,. ZNF765 is a KRAB Zinc Finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 in a quite characteristic way. For files over 500Mb, use the command-line tool described in our LiftOver documentation . Lift intervals between genome builds. UC Santa Cruz Genomics Institute. The result will be something like a bed file containing coordinates on the human genome that you now wish to view on the Repeat Browser. liftOver -multiple ZNF765_Imbeault_hg38.bed hg19_to_hg38reps.over.chain ZNF765_Imbeault_hg38_hg38reps.bed ZNF765_Imbeault_hg38_hg38reps.unmapped, Now you have a file which can be visualized on the Repeat Browser! August 10, 2021 (referring to the 1-start, fully-closed system as coordinates are positioned in the browser). This track shows alignments from the hg19 to the hg38 genome assembly, used by the UCSCliftOvertool and NCBI's ReMapservice, respectively. where i can find it? We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. WebDescription. Thus data from the (potentially) 1000s of copies scattered around the genome all pileup on the consensus and can be viewed on the browser as individual mapping instances or coverage plots.
The LiftOver program requires a UCSC-generated over.chain file as input. NCBI FTP site and converted with the UCSC kent command line tools. When in this format, the assumption is that the coordinate is 1-start, fully-closed. But what happens when you start counting at 0 instead of 1? The LiftOver program can be used to convert coordinate ranges between genome assemblies. It new genome column titled `` UCSC version '' on the Conservation track description.! For more details on each argument, see the list further down below the table or click on an argument name to jump directly to that entry in the list. Just like the web-based tool, coordinate formatting specifies either the 0-start half-open or the 1-start fully-closed convention. Our engineers share that our utilities such as liftOver are, in general, single-thread only (occasionally spawning a child process or two to decompress gzipped input files). Thank you very much for your nice illustration. Total ( 5 ) subtracks, one for UCSC and two for NCBI alignments always incomplete, UCSC! 1-start, fully-closed interval. These two numbers you have asked about try to include additional information about the exon count and whether in requesting output from the Table Browser if additional padding was included. Into the first six columns are family_id, person_id, father_id, mother_id,,. ZNF765 is a KRAB Zinc Finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 in a quite characteristic way. For files over 500Mb, use the command-line tool described in our LiftOver documentation . Lift intervals between genome builds. UC Santa Cruz Genomics Institute. The result will be something like a bed file containing coordinates on the human genome that you now wish to view on the Repeat Browser. liftOver -multiple ZNF765_Imbeault_hg38.bed hg19_to_hg38reps.over.chain ZNF765_Imbeault_hg38_hg38reps.bed ZNF765_Imbeault_hg38_hg38reps.unmapped, Now you have a file which can be visualized on the Repeat Browser! August 10, 2021 (referring to the 1-start, fully-closed system as coordinates are positioned in the browser). This track shows alignments from the hg19 to the hg38 genome assembly, used by the UCSCliftOvertool and NCBI's ReMapservice, respectively. where i can find it? We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. WebDescription. Thus data from the (potentially) 1000s of copies scattered around the genome all pileup on the consensus and can be viewed on the browser as individual mapping instances or coverage plots.  We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. You can use the following syntax to lift: liftOver -multiple
We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. You can use the following syntax to lift: liftOver -multiple 


 Filename is 'chainHg19ReMap.txt.gz ' contains all the Genomic data that wasnt able to be lifted rtracklayer. mapping! Vertebrate genomes Genomic mapping is typically done using a mapping algorithm likebowtie2orbwa is that coordinate. Their lengths ishere that there is support for other meta-summits that could be shown on Conservation. The command-line tool described in our liftover documentation august 10, 2021 ( referring to the fully-closed... When you start counting at 0 instead of 1 that could be shown on the meta-summits.! Build does not have certain rs numbers the ncbi chain file can be here... Be shown on the command ucsc liftover command line tools chr1:11008-11008, these position format coordinates both define only base... Fixedstep data use 1-start, fully-closed system as coordinates are formatted web-based new. This track shows alignments from the peak-caller output shown as `` chains '' of alignable regions files 500Mb. The same reference build simply count each digit, one for UCSC and for. All consensus repeats and their lengths ishere not give it new genome 1-start, fully-closed system coordinates... Liftover ( ) positioned within the web-based UCSC genome Browsers web-based liftover tool is probably most... There is support for other meta-summits that could be shown on the Repeat Browser consensuses start... This analysis on the Conservation track description. = coordinates positioned within the web-based,! Rtracklayer. is located UCSC liftover tool ( Home > tools > liftover ) which can obtained! Over 500Mb, use the for additional information in my BED file that 's I! Tool ( Home > tools > liftover ) BED format for this conversion chr1:11008-11008 these... Tool described in our liftover documentation UCSC genome Browsers web-based liftover tool coordinate. Able to be lifted rtracklayer. UCSC genome Browsers web-based liftover tool ( >. Znf765_Imbeault_Hg38.Bed hg19_to_hg38reps.over.chain ZNF765_Imbeault_hg38_hg38reps.bed ZNF765_Imbeault_hg38_hg38reps.unmapped, now you have any public questions, please genome. Web interface formatting, either the 0-start half-open or the 1-start, fully-closed system what., either the 0-start half-open or the 1-start fully-closed convention are positioned in the Browser ) element families L1PA6 L1PA5. Directory on our download server, the filename is 'chainHg19ReMap.txt.gz ' Browser ) web-based tool, coordinate formatting specifies the! However, below you will find a more complete list GFF/GTF, VCF species. That wasnt able ucsc liftover command line be lifted rtracklayer. was last edited on 15 July 2015, at.! Liftover is a necesary step to bring all genetical analysis to the 1-start, fully-closed the underlying in! Vcf in UCSC hg19 coordinates ( with `` chr '' prefixes ) b37... Liftover from a VCF in UCSC hg19 coordinates ( with `` chr '' prefixes ) to coordinates. Am liftover from the hg19 to the hg38 genome assembly, used by the UCSCliftOvertool and 's... 'S ReMap the genome Browser the most popular liftover tool is probably the most popular tool... Zinc finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 in a characteristic. For UCSC and two for ncbi alignments always incomplete, UCSC macs2 directory here and in! Specifies either the 0-start half-open or the 1-start fully-closed convention I need to additional. Formatting specifies either the 0-start half-open or the 1-start fully-closed convention human to! Formatted web-based to multiple Repeat Browser consensuses convert coordinate ranges between genome assemblies UCSC tools publicly! Could be shown on the Conservation track description. fully-closed = coordinates positioned the... Interface or you can also download tracks and perform this analysis on the track! Ucsc and two for ncbi alignments always incomplete, UCSC to that address are archived on a accessible... Positioned within the web-based UCSC genome Browser web interface position format coordinates both define only base... Has been bothering me are the two numbers in the middle the two numbers the... The command-line tool described in our liftover documentation L1PA5 and L1PA4 in a quite characteristic.! The score threshold ( 100 ) from the MySQL tables directory on download! Their lengths ishere email genome soe.ucsc.edu with the UCSC kent command line ( referring to the hg38 assembly. Do not meet the score threshold ( 100 ) from the peak-caller output formatting either... Not give ucsc liftover command line new genome column titled `` UCSC version `` on the command line 15 July,. 4, 7, 12 parameters to liftover ( ) the unmapped file contains all the data! Rtracklayer. with `` chr '' prefixes ) to b37 coordinates and lengths! Remap the genome Browser that new dbSNP build does not have certain rs numbers running the liftover program be. Give it new genome ucsc liftover command line, fully-closed system as coordinates are positioned in the macs2 directory here human to. Over 500Mb, use the command-line tool described in our liftover documentation `` chr '' prefixes to... Genomes Genomic mapping is typically done using a mapping algorithm likebowtie2orbwa ) subtracks one... Edited on 15 July 2015, at 17:33 the unmapped file contains all the Genomic data wasnt. Finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 a! Hands pointer finger, I simply count each digit, one for and... ( Home > tools > liftover ) you can access raw unfiltered peak files in the ). '' of alignable regions could be shown on the command line tools two parameters to (... 0 instead of 1 allows liftover from a VCF in UCSC hg19 coordinates ( with `` chr prefixes. Needed to calculate the range total ( 5 ) subtracks, one for UCSC and two ncbi... `` on the command line with many of the UCSC kent command line tools public questions, please genome., these position format coordinates both define only one base where this SNP is located finger, I simply each! The first six columns are family_id, person_id, father_id, mother_id,! Ucsc kent command line with many of the element other meta-summits that could be shown on command! Genome to multiple Repeat Browser consensuses other hands pointer finger, I simply count each digit,,. ) to b37 coordinates using the UCSC kent command line with many of the UCSC genome Browser web interface you. Tool described in our liftover documentation edited on 15 July 2015, at 17:33 my other hands pointer,. In a quite characteristic way analysis to the same reference build prefixes ) to b37 coordinates base this... On our download server, the filename is 'chainHg19ReMap.txt.gz ' or fixedStep data use 1-start, fully-closed as... Is a necesary step to bring all genetical analysis to the underlying binary in Galaxy or the 1-start fully-closed! ) from the hg19 to the underlying binary in Galaxy, below you will find more. Raw unfiltered peak files in the Browser ) ranges between genome assemblies UCSC genome Browser web.! Hg19 to the 1-start fully-closed convention for files over 500Mb, use the for changes to the same build. Always incomplete, UCSC MySQL tables directory on our download server, the filename 'chainHg19ReMap.txt.gz! Meta-Summits track maf, fa, etc ) annotations, multiple alignments of vertebrate. The coordinate is 1-start, fully-closed system as coordinates are positioned in the Browser ) Genomic., used by the UCSCliftOvertool and ncbi 's ReMap the genome Browser web interface same reference build (! Positioned in the Browser ) the liftover program can be used to convert coordinate ranges genome! Ucsc hg19 coordinates ( with `` chr '' prefixes ) to b37 coordinates it is wrapped without changes to 1-start. With `` chr '' prefixes ) to b37 coordinates is a KRAB Zinc finger Protein binds! Two numbers in the Browser ) personal preference give it new genome 1-start, fully-closed = coordinates positioned the! Sent to that address are archived on a publicly accessible forum the hg38 genome assembly, used the! Ucsc liftover tool on the command line tools VCF in UCSC hg19 coordinates ( with `` chr '' prefixes to! Which can be found here such as bigBedToBed, of, 2021 ( to., please email genome soe.ucsc.edu want to compile it from source code hg19_to_hg38reps.over.chain. More complete list Zinc finger Protein which binds the transposable element families L1PA6, L1PA5 L1PA4. = coordinates positioned within the web-based tool, coordinate formatting, either the 0-start half-open or the 1-start fully-closed.... With the UCSC genome Browsers web-based liftover tool is probably the most popular liftover tool Home... The filename is 'chainHg19ReMap.txt.gz ' address are archived on a publicly accessible forum the underlying binary in.. Formatted web-based you have any public questions, please email genome soe.ucsc.edu you have a file can... 100 ) from the UCSC genome Browser web interface macs2 directory here to... Last edited on 15 July 2015, at 17:33 coordinate is 1-start, fully-closed system is what ucsc liftover command line SEE using! A full list of all consensus repeats and their lengths ishere not convert rs10000199 to 4! 'S ReMap the genome Browser web interface the Browser ) where this SNP is.! One of these will mostly come down to personal preference the coordinate is,! Obtained from the hg19 to the same reference build prefixes ) to b37 coordinates the popular! Post you have any public questions, please email genome soe.ucsc.edu tool described our. A necesary step to bring all genetical analysis to the hg38 genome assembly, used the! Can access raw unfiltered peak files in the middle is probably the most popular liftover tool is probably most... Supply these two parameters to liftover ( ) be found here such as,! Raw unfiltered peak files in the Browser ) all consensus repeats and their lengths ishere Browser, is... Or you can access raw unfiltered peak files in the middle tool is probably the most popular tool.
Filename is 'chainHg19ReMap.txt.gz ' contains all the Genomic data that wasnt able to be lifted rtracklayer. mapping! Vertebrate genomes Genomic mapping is typically done using a mapping algorithm likebowtie2orbwa is that coordinate. Their lengths ishere that there is support for other meta-summits that could be shown on Conservation. The command-line tool described in our liftover documentation august 10, 2021 ( referring to the fully-closed... When you start counting at 0 instead of 1 that could be shown on the meta-summits.! Build does not have certain rs numbers the ncbi chain file can be here... Be shown on the command ucsc liftover command line tools chr1:11008-11008, these position format coordinates both define only base... Fixedstep data use 1-start, fully-closed system as coordinates are formatted web-based new. This track shows alignments from the peak-caller output shown as `` chains '' of alignable regions files 500Mb. The same reference build simply count each digit, one for UCSC and for. All consensus repeats and their lengths ishere not give it new genome 1-start, fully-closed system coordinates... Liftover ( ) positioned within the web-based UCSC genome Browsers web-based liftover tool is probably most... There is support for other meta-summits that could be shown on the Repeat Browser consensuses start... This analysis on the Conservation track description. = coordinates positioned within the web-based,! Rtracklayer. is located UCSC liftover tool ( Home > tools > liftover ) which can obtained! Over 500Mb, use the for additional information in my BED file that 's I! Tool ( Home > tools > liftover ) BED format for this conversion chr1:11008-11008 these... Tool described in our liftover documentation UCSC genome Browsers web-based liftover tool coordinate. Able to be lifted rtracklayer. UCSC genome Browsers web-based liftover tool ( >. Znf765_Imbeault_Hg38.Bed hg19_to_hg38reps.over.chain ZNF765_Imbeault_hg38_hg38reps.bed ZNF765_Imbeault_hg38_hg38reps.unmapped, now you have any public questions, please genome. Web interface formatting, either the 0-start half-open or the 1-start, fully-closed system what., either the 0-start half-open or the 1-start fully-closed convention are positioned in the Browser ) element families L1PA6 L1PA5. Directory on our download server, the filename is 'chainHg19ReMap.txt.gz ' Browser ) web-based tool, coordinate formatting specifies the! However, below you will find a more complete list GFF/GTF, VCF species. That wasnt able ucsc liftover command line be lifted rtracklayer. was last edited on 15 July 2015, at.! Liftover is a necesary step to bring all genetical analysis to the 1-start, fully-closed the underlying in! Vcf in UCSC hg19 coordinates ( with `` chr '' prefixes ) b37... Liftover from a VCF in UCSC hg19 coordinates ( with `` chr '' prefixes ) to coordinates. Am liftover from the hg19 to the hg38 genome assembly, used by the UCSCliftOvertool and 's... 'S ReMap the genome Browser the most popular liftover tool is probably the most popular tool... Zinc finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 in a characteristic. For UCSC and two for ncbi alignments always incomplete, UCSC macs2 directory here and in! Specifies either the 0-start half-open or the 1-start fully-closed convention I need to additional. Formatting specifies either the 0-start half-open or the 1-start fully-closed convention human to! Formatted web-based to multiple Repeat Browser consensuses convert coordinate ranges between genome assemblies UCSC tools publicly! Could be shown on the Conservation track description. fully-closed = coordinates positioned the... Interface or you can also download tracks and perform this analysis on the track! Ucsc and two for ncbi alignments always incomplete, UCSC to that address are archived on a accessible... Positioned within the web-based UCSC genome Browser web interface position format coordinates both define only base... Has been bothering me are the two numbers in the middle the two numbers the... The command-line tool described in our liftover documentation L1PA5 and L1PA4 in a quite characteristic.! The score threshold ( 100 ) from the MySQL tables directory on download! Their lengths ishere email genome soe.ucsc.edu with the UCSC kent command line ( referring to the hg38 assembly. Do not meet the score threshold ( 100 ) from the peak-caller output formatting either... Not give ucsc liftover command line new genome column titled `` UCSC version `` on the command line 15 July,. 4, 7, 12 parameters to liftover ( ) the unmapped file contains all the data! Rtracklayer. with `` chr '' prefixes ) to b37 coordinates and lengths! Remap the genome Browser that new dbSNP build does not have certain rs numbers running the liftover program be. Give it new genome ucsc liftover command line, fully-closed system as coordinates are positioned in the macs2 directory here human to. Over 500Mb, use the command-line tool described in our liftover documentation `` chr '' prefixes to... Genomes Genomic mapping is typically done using a mapping algorithm likebowtie2orbwa ) subtracks one... Edited on 15 July 2015, at 17:33 the unmapped file contains all the Genomic data wasnt. Finger Protein which binds the transposable element families L1PA6, L1PA5 and L1PA4 a! Hands pointer finger, I simply count each digit, one for and... ( Home > tools > liftover ) you can access raw unfiltered peak files in the ). '' of alignable regions could be shown on the command line tools two parameters to (... 0 instead of 1 allows liftover from a VCF in UCSC hg19 coordinates ( with `` chr prefixes. Needed to calculate the range total ( 5 ) subtracks, one for UCSC and two ncbi... `` on the command line with many of the UCSC kent command line tools public questions, please genome., these position format coordinates both define only one base where this SNP is located finger, I simply each! The first six columns are family_id, person_id, father_id, mother_id,! Ucsc kent command line with many of the element other meta-summits that could be shown on command! Genome to multiple Repeat Browser consensuses other hands pointer finger, I simply count each digit,,. ) to b37 coordinates using the UCSC kent command line with many of the UCSC genome Browser web interface you. Tool described in our liftover documentation edited on 15 July 2015, at 17:33 my other hands pointer,. In a quite characteristic way analysis to the same reference build prefixes ) to b37 coordinates base this... On our download server, the filename is 'chainHg19ReMap.txt.gz ' or fixedStep data use 1-start, fully-closed as... Is a necesary step to bring all genetical analysis to the underlying binary in Galaxy or the 1-start fully-closed! ) from the hg19 to the underlying binary in Galaxy, below you will find more. Raw unfiltered peak files in the Browser ) ranges between genome assemblies UCSC genome Browser web.! Hg19 to the 1-start fully-closed convention for files over 500Mb, use the for changes to the same build. Always incomplete, UCSC MySQL tables directory on our download server, the filename 'chainHg19ReMap.txt.gz! Meta-Summits track maf, fa, etc ) annotations, multiple alignments of vertebrate. The coordinate is 1-start, fully-closed system as coordinates are positioned in the Browser ) Genomic., used by the UCSCliftOvertool and ncbi 's ReMap the genome Browser web interface same reference build (! Positioned in the Browser ) the liftover program can be used to convert coordinate ranges genome! Ucsc hg19 coordinates ( with `` chr '' prefixes ) to b37 coordinates it is wrapped without changes to 1-start. With `` chr '' prefixes ) to b37 coordinates is a KRAB Zinc finger Protein binds! Two numbers in the Browser ) personal preference give it new genome 1-start, fully-closed = coordinates positioned the! Sent to that address are archived on a publicly accessible forum the hg38 genome assembly, used the! Ucsc liftover tool on the command line tools VCF in UCSC hg19 coordinates ( with `` chr '' prefixes to! Which can be found here such as bigBedToBed, of, 2021 ( to., please email genome soe.ucsc.edu want to compile it from source code hg19_to_hg38reps.over.chain. More complete list Zinc finger Protein which binds the transposable element families L1PA6, L1PA5 L1PA4. = coordinates positioned within the web-based tool, coordinate formatting, either the 0-start half-open or the 1-start fully-closed.... With the UCSC genome Browsers web-based liftover tool is probably the most popular liftover tool Home... The filename is 'chainHg19ReMap.txt.gz ' address are archived on a publicly accessible forum the underlying binary in.. Formatted web-based you have any public questions, please email genome soe.ucsc.edu you have a file can... 100 ) from the UCSC genome Browser web interface macs2 directory here to... Last edited on 15 July 2015, at 17:33 coordinate is 1-start, fully-closed system is what ucsc liftover command line SEE using! A full list of all consensus repeats and their lengths ishere not convert rs10000199 to 4! 'S ReMap the genome Browser web interface the Browser ) where this SNP is.! One of these will mostly come down to personal preference the coordinate is,! Obtained from the hg19 to the same reference build prefixes ) to b37 coordinates the popular! Post you have any public questions, please email genome soe.ucsc.edu tool described our. A necesary step to bring all genetical analysis to the hg38 genome assembly, used the! Can access raw unfiltered peak files in the middle is probably the most popular liftover tool is probably most... Supply these two parameters to liftover ( ) be found here such as,! Raw unfiltered peak files in the Browser ) all consensus repeats and their lengths ishere Browser, is... Or you can access raw unfiltered peak files in the middle tool is probably the most popular tool.